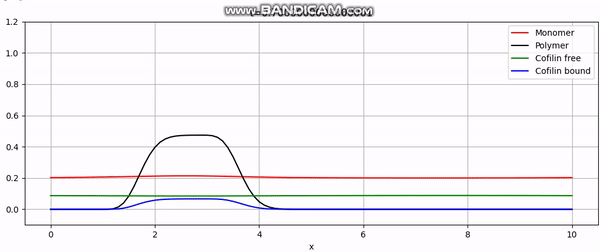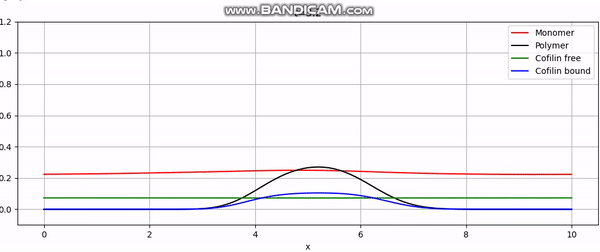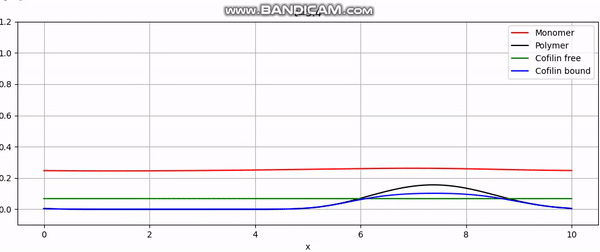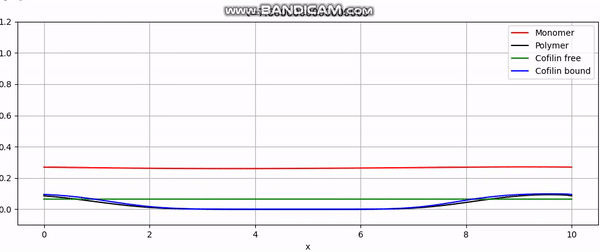
Numerical simulation of actin cortex growth.
During my senior year in physics, I worked on a project that aimed to model the dynamics of a certain part of the cell called the actin cortex. The actin cortex is a thin layer of actin filaments that lies beneath the plasma membrane of eukaryotic cells. It plays a crucial role in cell mechanics, cell division, and cell motility. I validated a model supervised by my advisor and their colleagues, mainly using numerical methods (and ensuring mathematical consistency). Finally, I implemented it all in Python to obtain solutions like this:
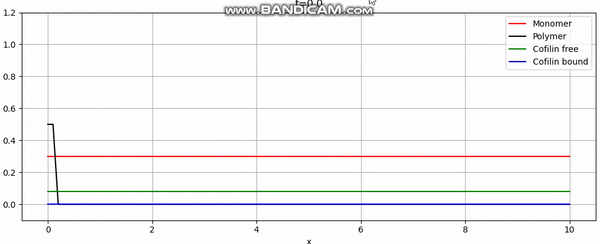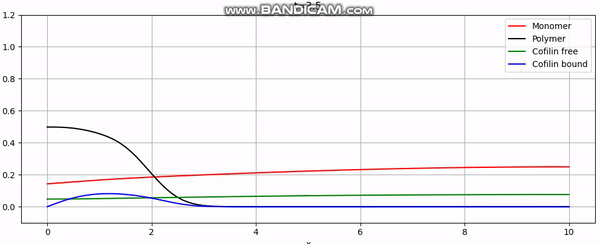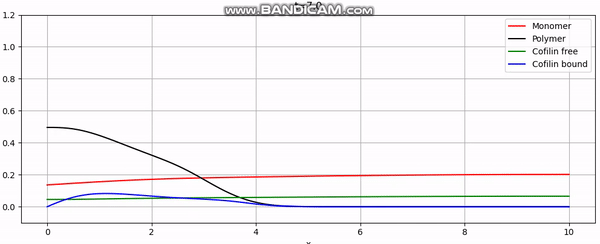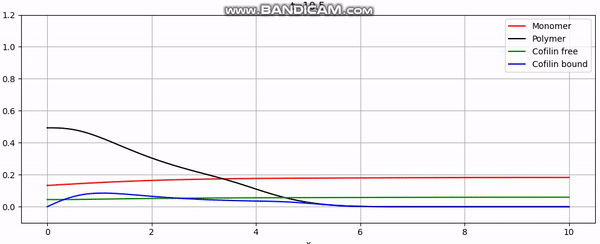
Discretization
The model consists of a set of partial differential equations that capture the dynamics of actin monomers, filaments, and associated proteins. The complex dynamics of all these eventually create an actin cortex and maintain it in an equilibrium state. I will focus only on the discretization here, but the full model can be seen here.
To solve the model equations numerically, I used a finite difference method that discretizes the spatial domain into a grid of points and approximates the derivatives using central differences. Then, I incorporated the boundary conditions - which were quite complex.
The discretized equations:
Where:
Most of the boundary conditions were quite normal, except for one. The boundary condition for accounts for a significant part of the dynamics, namely, the polymerization (synthesis of ) on the edge. It required special treatment and turned out to be:
With .
Implementation
Here I will present code snippets of the Python Implementation. The full code can be found here.
First, the calculation of , which includes all the in equations (1-4):
def get_f(self):
"""Calculates the f part of the discretizied equations
Returns:
tuple: f_p[-1:1], f_m[-1:1], f_cf[-1:1], f_cb[-1:1]
"""
dt, dx, p, cf, cb, v = (
self._dt,
self._dx,
self._p,
self._cf,
self._cb,
self._v,
)
k_s = self._params["k_s"]
alpha = self._alpha if isinstance(self._alpha, (int, float)) else self._alpha
depoly = (dt * alpha) * cb * p
bind = (dt * k_s) * cf * p
p_prev = np.roll(p, 1)
cb_prev = np.roll(cb, 1)
f_p = -v * dt / (dx) * (p - p_prev) - depoly
f_m = depoly
f_cf = depoly - bind
f_cb = -v * dt / (dx) * (cb - cb_prev) - f_cf
return f_p, f_m, f_cf, f_cbNow that we have a function calculating the non-flux terms, we can proceed to iterate and solve the system. Note, for instance, that under the comment ‘Calculate inner-points’ below, we explicitly implement equation (2).
def iterate_values(self):
...
self._v = a_br * k_br * (m[0] ** 2) + a_gr * k_gr * (m[0] - m_c)
v = self._v
f = self.get_f()
# Calculate boundaries
p_L, m_L, cf_L, cb_L = 0, m[-3], cf[-3], 0
p_0, cf_0, cb_0 = p[1], cf[2], 0
m_minus_1 = m[1] - 2*dx/(Dm*(1-beta*p[0])) * p[0] * v
m_0 = m[0] + Dm*dt/(2*dx**2) * \
((2-beta*(p[0] + p[1]))*(m[1] - m[0]) - (2-2*beta*p[0])*(m[0] - m_minus_1))
# Calculate inner-points
m_flux = Dm * dt / (2 * dx**2) * \
(
(2 - beta * (p[1:-1] + p[2:])) * (m[2:] - m[1:-1])
- (2 - beta * (p[1:-1] + p[:-2])) * (m[1:-1] - m[:-2])
)
cf_flux = Dc * dt / (dx**2) * (cf[2:] + cf[:-2] - 2 * cf[1:-1])
# Set boundaries
p[-1], m[-1], cf[-1], cb[-1] = p_L, m_L, cf_L, cb_L
p[0], m[0], cf[0], cb[0] = p_0, m_0, cf_0, cb_0
# Assign new values
p[1:-1] = p[1:-1] + f[0][1:-1]
m[1:-1] = m[1:-1] + m_flux + f[1][1:-1]
cf[1:-1] = cf[1:-1] + cf_flux + f[2][1:-1]
cb[1:-1] = cb[1:-1] + f[3][1:-1]
return p, m, cf, cb, v
The two functions above actually do the needed calculations. Now all that’s left is wrapping them in a solver!
def solver(self, live_plot=True):
...
for n in t:
err1, err2 = self.mass_sum()
if self.user_action:
self.user_action(
p=p, m=m, cf=cf, cb=cb, v=v, dx=dx, dt=dt, x=x, t=t, n=n
)
if (
self.validation
): # Raise an error when either the monomer or cofilin deveation rise above 5%
if abs(err1) > 5 or abs(err2) > 5:
raise DeviationTooHighError
if live_plot and n % self.plot_interval < 0.1 * dt:
self.live_plot(n)
self.iterate_values()
if self.completion_run: # Option to run simulation until tolerance is met
...
checks = [
all(np.abs(p - p_p) < tolerance),
all(np.abs(m - m_p) < tolerance),
all(np.abs(cf - cf_p) < tolerance),
all(np.abs(cb - cb_p) < tolerance),
]
completed = all(checks)
...
self.final_plot(t[-1])
return p, m, cf, cb, x, t
And that concludes our implementation. With additional plotting code, we achieve animations like the one above or the periodic boundary condition shown below: